The Unseen Regulator
An advanced exploration into the critical post-transcriptional control mechanisms governed by the mRNA 3'-UTR.
Learn More 👇 Methods 🔬Dive in with Flashcard Learning!
🎮 Play the Wiki2Web Clarity Challenge Game🎮
Introduction
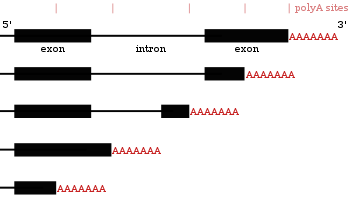
Defining the 3'-UTR
In molecular genetics, the three prime untranslated region (3'-UTR) denotes the segment of messenger RNA (mRNA) situated immediately following the translation termination codon. This region, while not coding for protein, frequently harbors regulatory sequences that exert significant post-transcriptional control over gene expression.
The Central Role in Gene Expression
Following transcription from DNA, mRNA molecules undergo translation into proteins. However, several regions of the mRNA, including the 5' cap, 5' UTR, 3' UTR, and poly(A) tail, are not translated. The 3'-UTR is particularly crucial, containing regulatory elements that modulate mRNA polyadenylation, translation efficiency, subcellular localization, and overall stability. It serves as a critical nexus for fine-tuning the cellular proteome.
Information Flow Context
The process of gene expression involves DNA being transcribed into RNA, which is then translated into protein. The 3'-UTR resides on the mRNA molecule after the stop codon, acting as a key regulatory element that influences how much protein is ultimately produced from that mRNA transcript. Its influence extends beyond mere stability to active participation in translational control.
Physical Characteristics
Variable Length
The 3'-UTR exhibits considerable variation in length across the mammalian genome. While some can be as short as 60 nucleotides, others extend up to approximately 4000 nucleotides. In humans, the average length is around 800 nucleotides, significantly longer than the average 5'-UTR length of approximately 200 nucleotides. This length disparity is functionally significant, as longer 3'-UTRs are often associated with lower levels of gene expression, potentially due to an increased probability of containing miRNA binding sites.
Nucleotide Composition
The nucleotide composition of UTRs differs notably between the 5' and 3' ends. In warm-blooded vertebrates, the 5'-UTR typically has a higher Guanine-Cytosine (G+C) content (around 60%) compared to the 3'-UTR (around 45%). A correlation exists where GC-poor UTRs tend to be longer than their GC-rich counterparts, influencing secondary structure formation and regulatory interactions.
Stability and Decay
Specific sequences within the 3'-UTR directly influence mRNA stability and degradation rates. AU-rich elements (AREs), typically 50-150 base pairs containing multiple AUUUA pentanucleotides, are key regulators. Proteins binding to AREs can modulate mRNA decay or affect translation initiation, playing roles in crucial cellular processes like cell growth, differentiation, and response to stimuli.
Key Regulatory Elements
MicroRNA Response Elements (MREs)
Many 3'-UTRs contain MREs, specific sequences recognized by microRNAs (miRNAs). miRNAs are small non-coding RNAs that bind to mRNA, typically via partial base pairing with their seed sequence in the MRE. This binding often leads to translational repression or direct mRNA degradation, thereby downregulating gene expression.
AU-Rich Elements (AREs)
AREs are regulatory sequences found within the 3'-UTR, crucial for modulating the stability and decay rates of specific transcripts. They bind a variety of RNA-binding proteins (ARE-BPs) whose activity is context-dependent (tissue type, cellular signals). AREs are particularly prevalent in mRNAs encoding cytokines, growth factors, and transcription factors, linking them to inflammation, immunity, and cell proliferation.
The Poly(A) Tail
While technically added post-transcriptionally, the poly(A) tail is initiated by signals within the 3'-UTR (e.g., the AAUAAA sequence). It binds poly(A) binding proteins (PABPs), which are essential for mRNA export, stability, and translation. PABP interaction with factors at the 5' cap promotes mRNA circularization, enhancing translation efficiency and ribosome recycling. Removal of the poly(A) tail typically triggers mRNA degradation.
Iron Response Elements (IREs)
IREs are stem-loop structures found in the UTRs of mRNAs encoding proteins involved in iron metabolism. When intracellular iron levels are low, specific proteins bind to IREs, stabilizing the mRNA and promoting protein synthesis (e.g., transferrin receptor). Conversely, high iron levels lead to dissociation, often resulting in mRNA degradation.
Cytoplasmic Polyadenylation Elements (CPEs)
CPEs are AU-rich sequences in the 3'-UTR that regulate translation, particularly of maternal mRNAs during early development. Binding of CPE-binding protein (CPEB) to CPEs, often near the nuclear polyadenylation signal, influences polyadenylation status and translational activation or repression.
SECIS Element
The Selenocysteine Insertion Sequence (SECIS) element is a conserved stem-loop structure within the 3'-UTR. It directs the incorporation of the unusual amino acid selenocysteine into the polypeptide chain at a standard UGA stop codon, enabling the synthesis of selenoproteins.
Structural Dynamics
Secondary Structures
Beyond linear sequences, the three-dimensional folding of the 3'-UTR into secondary structures, such as stem-loops, is critical for function. These structures can serve as scaffolds for RNA-binding proteins and non-coding RNAs, influencing transcript expression. Protein factors can modulate this folding, thereby controlling regulatory interactions.
Human Complexity
Human transcripts exhibit particularly long 3'-UTRs compared to other mammals, reflecting a higher degree of complexity in gene regulation. This increased length provides more potential sites for regulatory interactions, allowing for finer control over gene expression tailored to specific cellular contexts.
Alternative Polyadenylation (APA)
Generating Isoforms
Alternative polyadenylation (APA) is a mechanism generating mRNA isoforms that differ solely in their 3'-UTR lengths. This occurs when a gene possesses multiple polyadenylation sites. APA significantly impacts gene expression by altering the presence of regulatory elements (like miRNA binding sites) within the 3'-UTR, affecting mRNA stability, cytoplasmic export, and translation efficiency.
Functional Significance
APA is utilized by approximately half of all human genes and is particularly important in complex organisms. It provides a sophisticated mechanism for modulating protein levels and localization without altering the protein sequence itself, contributing to cellular diversity and developmental regulation.
Methods of Study
Computational Approaches
Bioinformatics tools enable rapid analysis of vast sequence datasets. Computational methods can identify potential regulatory elements like AREs in a significant percentage of human 3'-UTRs and predict miRNA target sites in a majority of them. Sequence comparison helps uncover conserved motifs and functional similarities across different species.
Experimental Techniques
Experimental methods are essential for validating computational predictions and defining the precise function of 3'-UTR elements. Techniques like RNA-binding protein immunoprecipitation (RIP) and cross-linking followed by sequencing (CLIP) map protein binding sites. Site-directed mutagenesis allows researchers to investigate the impact of specific sequence alterations or structural changes on mRNA processing, localization, stability, and translation.
Transcript-Wide Analysis
Advanced techniques such as ribosome profiling provide a global view of translation efficiency across all transcripts. Combined with other high-throughput sequencing methods, these approaches help elucidate the complex interplay between cis-acting elements in the 3'-UTR and trans-acting factors (proteins and miRNAs), revealing novel regulatory mechanisms and their contribution to cellular function.
Implications in Disease
Mutation Consequences
Alterations within the 3'-UTR can have widespread effects, potentially dysregulating the expression of multiple genes. Mutations affecting binding sites for RNA-binding proteins or miRNAs can disrupt normal cellular processes. For instance, dysregulation of ARE-binding proteins has been implicated in tumorigenesis, leukemia, and developmental disorders.
Myotonic Dystrophy
A classic example is myotonic dystrophy, caused by an expanded number of CTG repeats within the 3'-UTR of the dystrophia myotonica protein kinase (DMPK) gene. This expansion leads to aberrant RNA structures and protein binding, disrupting gene regulation and causing the characteristic muscle weakness and wasting.
Hematological and Neurological Links
Dysfunctional ARE-binding proteins are linked to various hematological malignancies. Furthermore, mutations or altered expression related to 3'-UTR elements have been associated with conditions such as acute myeloid leukemia, alpha-thalassemia, neuroblastoma, and certain congenital disorders, highlighting the critical role of these regions in human health.
Future Directions
Uncharted Territory
Despite significant advances, the 3'-UTR remains a complex and relatively understudied domain. The intricate network of overlapping regulatory elements, alternative polyadenylation sites, and numerous binding proteins presents a challenge for complete functional elucidation. Identifying the specific roles of each element and their interacting factors requires continued investigation.
Technological Advancements
Emerging technologies, particularly deep-sequencing based methods like ribosome profiling, are providing unprecedented resolution into RNA regulation. Future research leveraging these tools will undoubtedly uncover more subtle regulatory nuances, identify novel cis-elements and trans-acting factors, and deepen our understanding of the 3'-UTR's pervasive influence on cellular function and disease pathogenesis.
Teacher's Corner
Edit and Print this course in the Wiki2Web Teacher Studio

Click here to open the "Three Prime Untranslated Region" Wiki2Web Studio curriculum kit
Use the free Wiki2web Studio to generate printable flashcards, worksheets, exams, and export your materials as a web page or an interactive game.
True or False?
Test Your Knowledge!
Gamer's Corner
Are you ready for the Wiki2Web Clarity Challenge?
Unlock the mystery image and prove your knowledge by earning trophies. This simple game is addictively fun and is a great way to learn!
Play now
References
References
Feedback & Support
To report an issue with this page, or to find out ways to support the mission, please click here.
Disclaimer
Important Notice
This page was generated by an Artificial Intelligence and is intended for informational and educational purposes only. The content is derived from publicly available data and may not be exhaustive or entirely up-to-date.
This is not medical or biological advice. The information provided herein is not a substitute for professional consultation, diagnosis, or treatment from qualified healthcare providers or researchers. Always seek the advice of a qualified professional with any questions you may have regarding a medical condition or biological process. Never disregard professional advice or delay in seeking it because of information obtained from this resource.
The creators of this page are not responsible for any errors or omissions, or for any actions taken based on the information provided.